PoweREST
Installation
In R or Rstudio
# install devtools if necessary
install.packages('devtools')
# install the PoweREST package
devtools::install_github('lanshui98/PoweREST')
# load package
library(PoweREST)
If you have problems installing the package, please try to install necessary packages yourself from CRAN.
install.packages(c("scam","Seurat","dplyr","plotly","resample","ggplot2","xgboost","magrittr","rayshader"))
Suggested packages for the tutorial:
install.packages(c("patchwork","boot","knitr","rmarkdown","fields","rayrender","tidyr"))
Introduction to the package
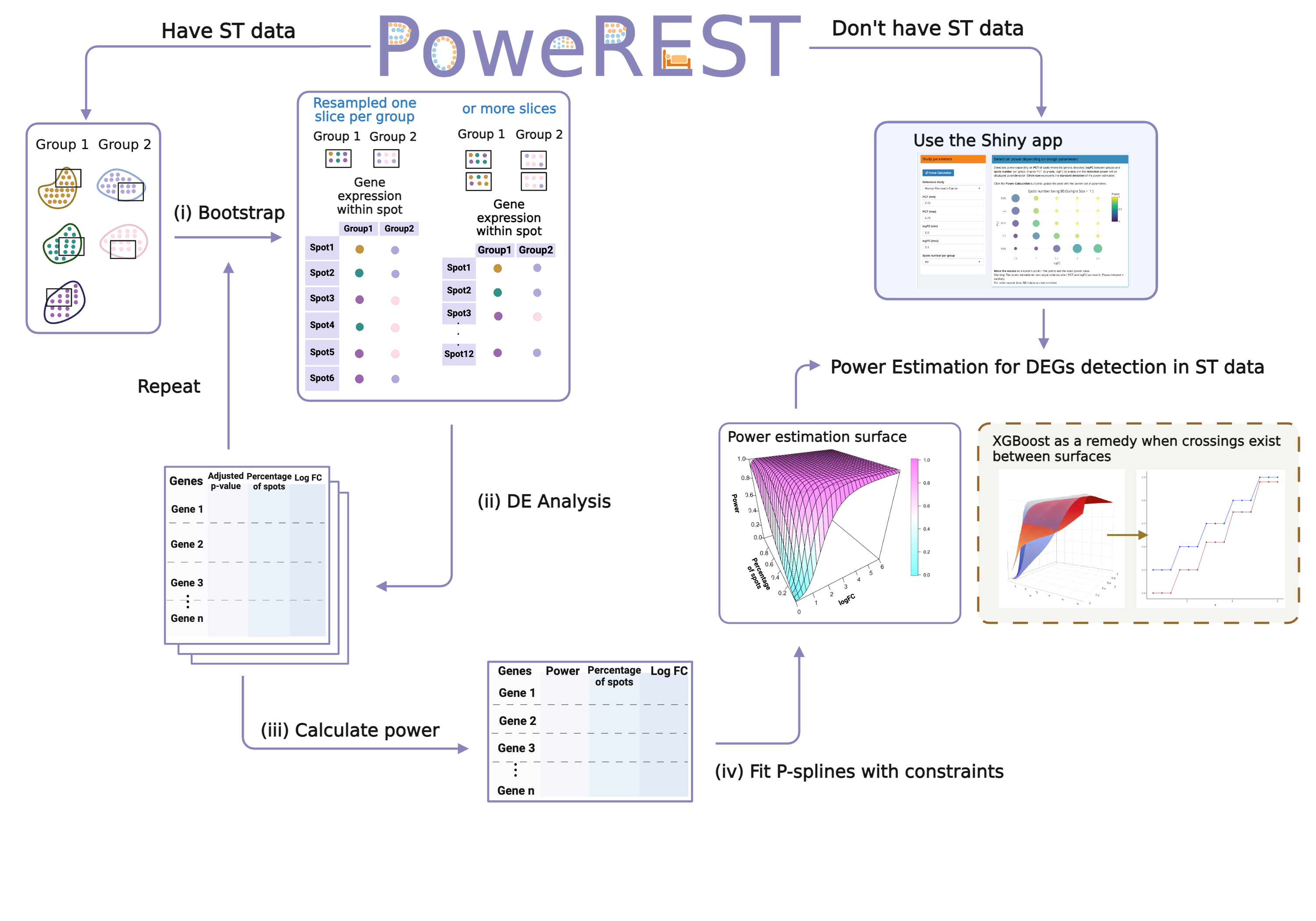
PoweREST is R package for the power analysis of detecting differential expressed genes between two conditions using 10X Visium spatial transcriptomics (ST). It enables the user to estimate the power or sample size needed for a 10X Visium ST experiment with and without prior dataset available by depicting how the study power is determined by three key parameters: (i) the number of biological replicates; (ii) the percentage of spots where the gene is detected in both groups; (iii) the log-fold change in average expression between two groups. PoweREST relies fully upon non-parametric modelling techniques but under biologically meaningful constraints which is extremely suitable for complex ST samples. The tool has been evaluated upon data from different tissue samples with promising and robust results.
Shiny app (no coding required)
A graphical interface of a shiny app is at the webpage PoweREST, which provides power estimations upon different pilot ST datasets.
Cite PoweREST
Shui, Lan, et al. “PoweREST: Statistical power estimation for spatial transcriptomics experiments to detect differentially expressed genes between two conditions.” PLOS Computational Biology 21.7 (2025): e1013293.
Contact us
If you have questions, feel free to leave messages on the github issues or contact me through email: lshui@mdanderson.org
Acknowledgement
Thanks for the guidance from Dr Ziyi Li and Dr Liang Li. This work was in part funded by the Translational and Basic Science Research in Early Lesions (TBEL) Program. This website is build based on CARD’s tutorial by Dr Ying Ma.